Anti-HR23B (GOAT) Antibody
HR23B Antibody
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND
| Host | Goat |
|---|---|
| Conjugate | Unconjugated |
| Target Species | Human |
| Reactivity | Human |
| Clonality | Polyclonal |
Application
| WB, E, I, LCI |
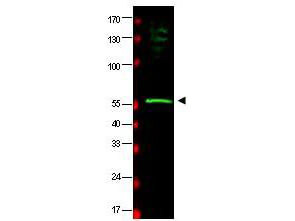
| Application Note | This affinity purified antibody has been tested for use in ELISA and by western blot. Specific conditions for reactivity should be optimized by the end user. Expect a band approximately 58 kDa in size corresponding to HR23B by western blotting in the appropriate cell lysate or extract. |
| Physical State | Liquid (sterile filtered) |
| Buffer | 0.02 M Potassium Phosphate, 0.15 M Sodium Chloride, pH 7.2 |
| Immunogen | This affinity purified antibody was prepared from whole goat serum produced by repeated immunizations with a synthetic peptide corresponding to an internal region near aa 155-180 of human HR23B protein. |
| Preservative | 0.01% (w/v) Sodium Azide |
| Gene ID | 5887 |
|---|---|
| Other Names | 5887 |
| Purity | This is an affinity-purified antibody produced by immunoaffinity chromatography using the immunizing peptide after immobilization to a solid phase. Reactivity occurs against human HR23B protein. Sequence homology as assessed by BLAST indicated 100% homology for this protein from human, dog, chimpanzee and S. cerevisiae. Cross reactivity with HR23B protein from mouse and rat may also occur as sequence homology varies by one amino acid residue in this sequence by BLAST analysis. Reactivity with HR23B protein from other sources is not known. |
| Storage Condition | Store vial at -20° C prior to opening. Aliquot contents and freeze at -20° C or below for extended storage. Avoid cycles of freezing and thawing. Centrifuge product if not completely clear after standing at room temperature. This product is stable for several weeks at 4° C as an undiluted liquid. Dilute only prior to immediate use. |
| Precautions Note | This product is for research use only and is not intended for therapeutic or diagnostic applications. |
| Name | RAD23B |
|---|---|
| Function | Multiubiquitin chain receptor involved in modulation of proteasomal degradation. Binds to polyubiquitin chains. Proposed to be capable to bind simultaneously to the 26S proteasome and to polyubiquitinated substrates and to deliver ubiquitinated proteins to the proteasome. May play a role in endoplasmic reticulum-associated degradation (ERAD) of misfolded glycoproteins by association with PNGase and delivering deglycosylated proteins to the proteasome. The XPC complex is proposed to represent the first factor bound at the sites of DNA damage and together with other core recognition factors, XPA, RPA and the TFIIH complex, is part of the pre-incision (or initial recognition) complex. The XPC complex recognizes a wide spectrum of damaged DNA characterized by distortions of the DNA helix such as single-stranded loops, mismatched bubbles or single-stranded overhangs. The orientation of XPC complex binding appears to be crucial for inducing a productive NER. XPC complex is proposed to recognize and to interact with unpaired bases on the undamaged DNA strand which is followed by recruitment of the TFIIH complex and subsequent scanning for lesions in the opposite strand in a 5'-to-3' direction by the NER machinery. Cyclobutane pyrimidine dimers (CPDs) which are formed upon UV-induced DNA damage esacpe detection by the XPC complex due to a low degree of structural perurbation. Instead they are detected by the UV-DDB complex which in turn recruits and cooperates with the XPC complex in the respective DNA repair. In vitro, the XPC:RAD23B dimer is sufficient to initiate NER; it preferentially binds to cisplatin and UV-damaged double-stranded DNA and also binds to a variety of chemically and structurally diverse DNA adducts. XPC:RAD23B contacts DNA both 5' and 3' of a cisplatin lesion with a preference for the 5' side. XPC:RAD23B induces a bend in DNA upon binding. XPC:RAD23B stimulates the activity of DNA glycosylases TDG and SMUG1. |
| Cellular Location | Nucleus. Cytoplasm. Note=The intracellular distribution is cell cycle dependent. Localized to the nucleus and the cytoplasm during G1 phase. Nuclear levels decrease during S-phase; upon entering mitosis, relocalizes in the cytoplasm without association with chromatin |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
HR23B (also known as UV excision repair protein RAD23 homolog B, XP-C repair complementing complex 58 kDa protein and p58) is one of two human homologs of Saccharomyces cerevisiae Rad23 (hHR23A and hHR23B), a protein involved in nucleotide excision repair (NER). This protein was shown to interact with, and elevate the nucleotide excision activity of 3-methyladenine-DNA glycosylase (MPG), which suggested a role in DNA damage recognition in base excision repair. This protein contains an N-terminal ubiquitin-like domain, which was reported to interact with 26S proteasome, as well as with ubiquitin protein ligase E6AP, and thus suggests that this protein may be involved in the ubiquitin mediated proteolytic pathway in cells.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.