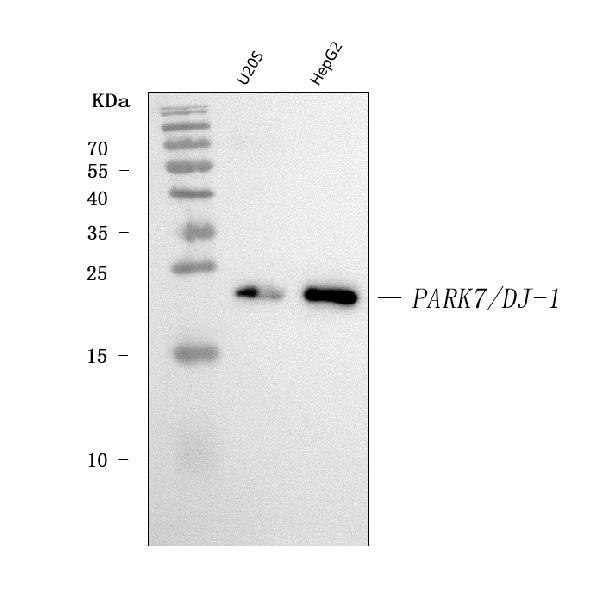
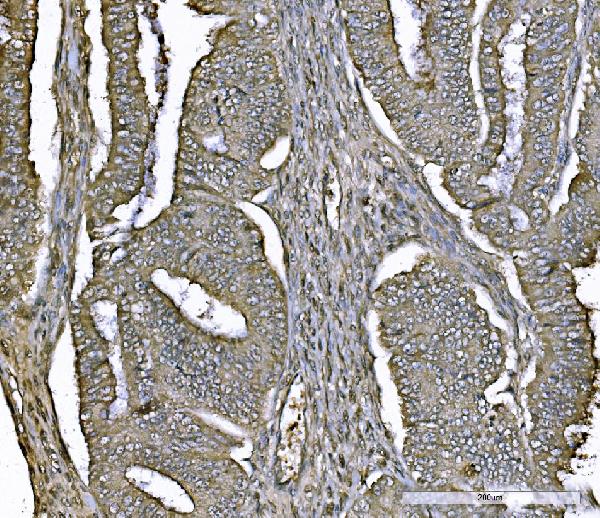
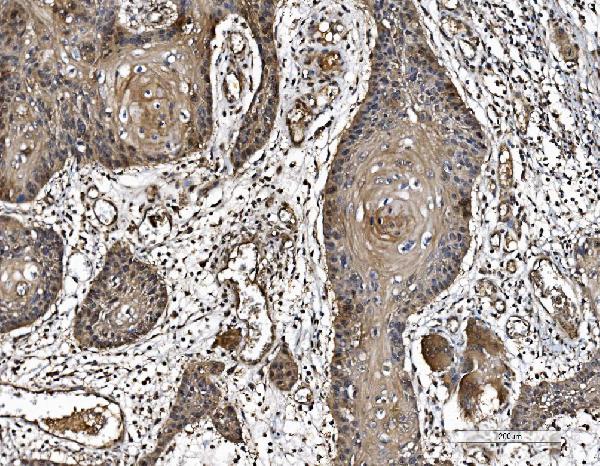
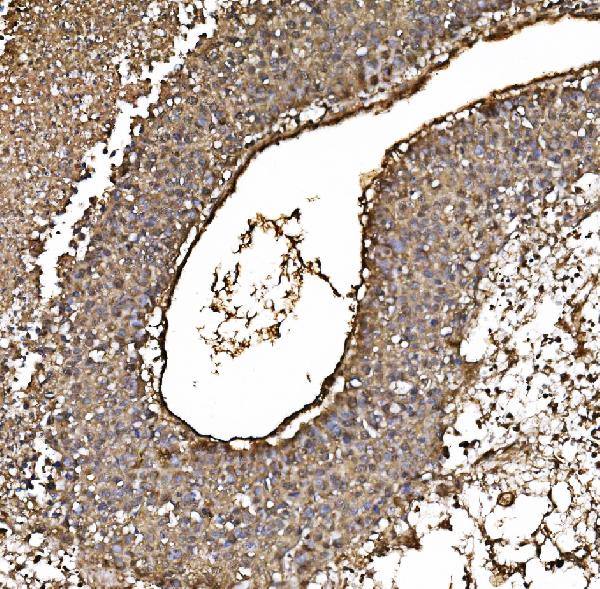
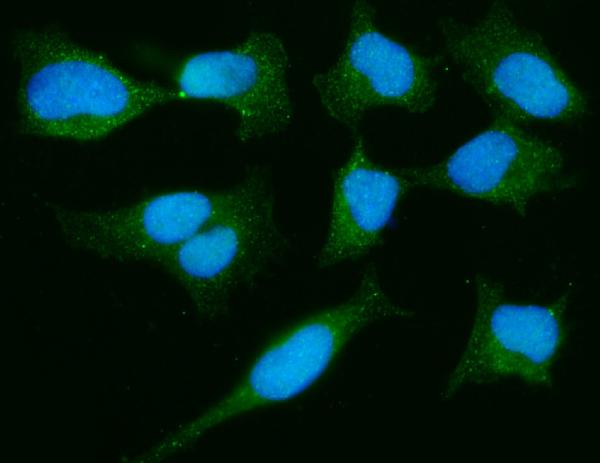
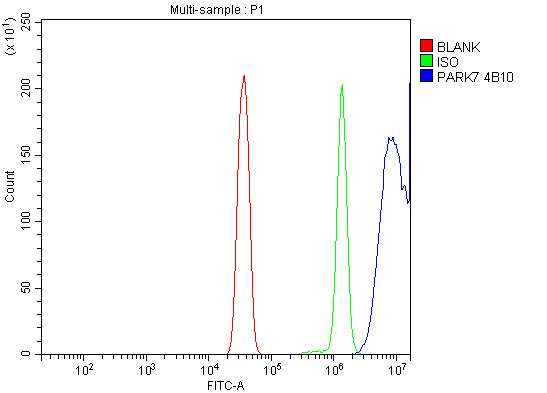
Anti-PARK7/DJ1 Antibody Picoband™ (monoclonal, 4B10)
- SPECIFICATION
- CITATIONS
- PROTOCOLS
- BACKGROUND

Application
| WB, IHC, IF, ICC, FC |
|---|---|
| Primary Accession | Q99497 |
| Host | Mouse |
| Isotype | IgG2b |
| Reactivity | Human |
| Clonality | Monoclonal |
| Format | Lyophilized |
| Description | Anti-PARK7/DJ1 Antibody Picoband™ (monoclonal, 4B10) . Tested in Flow Cytometry, IF, IHC, ICC, WB applications. This antibody reacts with Human. |
| Reconstitution | Adding 0.2 ml of distilled water will yield a concentration of 500 µg/ml. |
| Gene ID | 11315 |
|---|---|
| Other Names | Parkinson disease protein 7, Maillard deglycase, Oncogene DJ1, Parkinsonism-associated deglycase {ECO:0000312|HGNC:HGNC:16369}, Protein DJ-1, DJ-1, Protein/nucleic acid deglycase DJ-1, 3.1.2.-, 3.5.1.-, 3.5.1.124, PARK7 (HGNC:16369) |
| Calculated MW | 22 kDa |
| Application Details | Western blot, 0.25-0.5 µg/ml, Human Immunohistochemistry(Paraffin-embedded Section), 2-5 µg/ml, Human Immunocytochemistry/Immunofluorescence, 5 µg/ml, Human Flow Cytometry, 1-3 µg/1x10^6 cells, Human |
| Contents | Each vial contains 4 mg Trehalose, 0.9 mg NaCl and 0.2 mg Na2HPO4. |
| Clone Names | Clone: 4B10 |
| Immunogen | E.coli-derived human PARK7 recombinant protein (Position: A2-D189). Human PARK7 shares 91% amino acid (aa) sequence identity with both mouse and rat PARK7. |
| Purification | Immunogen affinity purified. |
| Storage | At -20°C for one year from date of receipt. After reconstitution, at 4°C for one month. It can also be aliquotted and stored frozen at -20°C for six months. Avoid repeated freezing and thawing. |
| Name | PARK7 (HGNC:16369) |
|---|---|
| Function | Multifunctional protein with controversial molecular function which plays an important role in cell protection against oxidative stress and cell death acting as oxidative stress sensor and redox- sensitive chaperone and protease (PubMed:12796482, PubMed:17015834, PubMed:18711745, PubMed:19229105, PubMed:20304780, PubMed:25416785, PubMed:26995087, PubMed:28993701). It is involved in neuroprotective mechanisms like the stabilization of NFE2L2 and PINK1 proteins, male fertility as a positive regulator of androgen signaling pathway as well as cell growth and transformation through, for instance, the modulation of NF-kappa-B signaling pathway (PubMed:12612053, PubMed:14749723, PubMed:15502874, PubMed:17015834, PubMed:18711745, PubMed:21097510). Has been described as a protein and nucleotide deglycase that catalyzes the deglycation of the Maillard adducts formed between amino groups of proteins or nucleotides and reactive carbonyl groups of glyoxals (PubMed:25416785, PubMed:28596309). But this function is rebuted by other works (PubMed:27903648, PubMed:31653696). As a protein deglycase, repairs methylglyoxal- and glyoxal-glycated proteins, and releases repaired proteins and lactate or glycolate, respectively. Deglycates cysteine, arginine and lysine residues in proteins, and thus reactivates these proteins by reversing glycation by glyoxals. Acts on early glycation intermediates (hemithioacetals and aminocarbinols), preventing the formation of advanced glycation endproducts (AGE) that cause irreversible damage (PubMed:25416785, PubMed:26995087, PubMed:28013050). Also functions as a nucleotide deglycase able to repair glycated guanine in the free nucleotide pool (GTP, GDP, GMP, dGTP) and in DNA and RNA. Is thus involved in a major nucleotide repair system named guanine glycation repair (GG repair), dedicated to reversing methylglyoxal and glyoxal damage via nucleotide sanitization and direct nucleic acid repair (PubMed:28596309). Protects histones from adduction by methylglyoxal, controls the levels of methylglyoxal- derived argininine modifications on chromatin (PubMed:30150385). Able to remove the glycations and restore histone 3, histone glycation disrupts both local and global chromatin architecture by altering histone-DNA interactions as well as histone acetylation and ubiquitination levels (PubMed:30150385, PubMed:30894531). Displays a very low glyoxalase activity that may reflect its deglycase activity (PubMed:22523093, PubMed:28993701, PubMed:31653696). Eliminates hydrogen peroxide and protects cells against hydrogen peroxide-induced cell death (PubMed:16390825). Required for correct mitochondrial morphology and function as well as for autophagy of dysfunctional mitochondria (PubMed:16632486, PubMed:19229105). Plays a role in regulating expression or stability of the mitochondrial uncoupling proteins SLC25A14 and SLC25A27 in dopaminergic neurons of the substantia nigra pars compacta and attenuates the oxidative stress induced by calcium entry into the neurons via L-type channels during pacemaking (PubMed:18711745). Regulates astrocyte inflammatory responses, may modulate lipid rafts-dependent endocytosis in astrocytes and neuronal cells (PubMed:23847046). In pancreatic islets, involved in the maintenance of mitochondrial reactive oxygen species (ROS) levels and glucose homeostasis in an age- and diet dependent manner. Protects pancreatic beta cells from cell death induced by inflammatory and cytotoxic setting (By similarity). Binds to a number of mRNAs containing multiple copies of GG or CC motifs and partially inhibits their translation but dissociates following oxidative stress (PubMed:18626009). Metal-binding protein able to bind copper as well as toxic mercury ions, enhances the cell protection mechanism against induced metal toxicity (PubMed:23792957). In macrophages, interacts with the NADPH oxidase subunit NCF1 to direct NADPH oxidase-dependent ROS production, and protects against sepsis (By similarity). |
| Cellular Location | Cell membrane {ECO:0000250|UniProtKB:Q99LX0}; Lipid-anchor {ECO:0000250|UniProtKB:Q99LX0}. Cytoplasm. Nucleus. Membrane raft {ECO:0000250|UniProtKB:O88767}. Mitochondrion. Endoplasmic reticulum. Note=Under normal conditions, located predominantly in the cytoplasm and, to a lesser extent, in the nucleus and mitochondrion. Translocates to the mitochondrion and subsequently to the nucleus in response to oxidative stress and exerts an increased cytoprotective effect against oxidative damage (PubMed:18711745). Detected in tau inclusions in brains from neurodegenerative disease patients (PubMed:14705119). Membrane raft localization in astrocytes and neuronal cells requires palmitoylation |
| Tissue Location | Highly expressed in pancreas, kidney, skeletal muscle, liver, testis and heart. Detected at slightly lower levels in placenta and brain (at protein level). Detected in astrocytes, Sertoli cells, spermatogonia, spermatids and spermatozoa. Expressed by pancreatic islets at higher levels than surrounding exocrine tissues (PubMed:22611253). |

Thousands of laboratories across the world have published research that depended on the performance of antibodies from Abcepta to advance their research. Check out links to articles that cite our products in major peer-reviewed journals, organized by research category.
info@abcepta.com, and receive a free "I Love Antibodies" mug.
Provided below are standard protocols that you may find useful for product applications.
Background
Parkinson disease (autosomal recessive, early onset) 7, also known as DJ1, is a protein which in humans is encoded by the PARK7 gene. PARK7 belongs to the peptidase C56 family of proteins. PARK7 is mapped to chromosome 1p36. It acts as a positive regulator of androgen receptor-dependent transcription. It is also involved in tumorigenesis and in maintaining mitochondrial homeostasis. This gene may also function as a redox-sensitive chaperone, as a sensor foroxidative stress, and it apparently protects neurons against oxidative stress and cell death. It has been found that PARK7 mutations that impair transcriptional coactivator function can render dopaminergic neurons vulnerable to apoptosis and may contribute to the pathogenesis of Parkinson disease.
If you have used an Abcepta product and would like to share how it has performed, please click on the "Submit Review" button and provide the requested information. Our staff will examine and post your review and contact you if needed.
If you have any additional inquiries please email technical services at tech@abcepta.com.













 Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them.
Foundational characteristics of cancer include proliferation, angiogenesis, migration, evasion of apoptosis, and cellular immortality. Find key markers for these cellular processes and antibodies to detect them. The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.
The SUMOplot™ Analysis Program predicts and scores sumoylation sites in your protein. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.
The Autophagy Receptor Motif Plotter predicts and scores autophagy receptor binding sites in your protein. Identifying proteins connected to this pathway is critical to understanding the role of autophagy in physiological as well as pathological processes such as development, differentiation, neurodegenerative diseases, stress, infection, and cancer.